phylogenomics

formins
In eukaryotes, the assembly of globular actin monomers into linear filaments is controlled by formin proteins. Formins are long molecules of >1,000 amino acid residues that are composed of various combinations of different functional domains. Formins are encoded by a multigene family, the size and content of which varies with organism.
We investigated the evolutionary relationships among different formin genes and their origin. To clarify the origin and evolution of the formin gene family and to infer the domain organization of the ancestral formin molecule we surveyed a large set of genomes and proteomes . Because formins contain various domain combinations, we have used these combinations as synapomorphies (derived character states shared by two or more taxa) to study the evolutionary relationships of the formin genes.
Our study on the formin gene family suggested that the formin homology 2 domain, which promotes the formation of actin filaments, is a eukaryotic innovation and originated only once in eukaryotic evolution. The formin genes have experienced multiple taxon‐specific duplications and followed the birth‐and‐death model of evolution. Additionally, they experienced taxon‐specific genomic rearrangements that led to the acquisition of unrelated protein domains. The evolutionary diversification of formin genes apparently increased the number of FORMINs interacting molecules and consequently contributed to the development of a complex and precise actin assembly mechanism. The diversity of FORMIN types is probably related to the range of actin‐based cellular processes that different cells or organisms require. Our results indicated the importance of gene duplication and domain acquisition in the evolution of the eukaryotic cell and offered insights into how a complex system, such as the cytoskeleton, evolved.

dangers
Developmental proteins play a pivotal role in the origin of animal complexity and diversity. We identified a highly divergent developmental protein superfamily, DANGER, which originated before the emergence of animals (∼850 million years ago) and experienced major expansion-contraction events during metazoan evolution. Our sequence analysis demonstrated that DANGER proteins diverged via multiple mechanisms, including amino acid substitution, intron gain and/or loss, and recombination. Divergence for DANGER proteins is substantially greater than for the prototypic member of the superfamily (Mab-21 family) and other developmental protein families (e.g., WNT proteins). We found that DANGER proteins are widely expressed, and display species-dependent tissue expression patterns, with many members having roles in development. We determined that DANGER1A, which regulates the inositol trisphosphate receptor, promotes the differentiation and outgrowth of neuronal processes. Regulation of development may be a universal function of DANGER family members. This family provides a model system to investigate how rapid protein divergence contributes to morphological complexity.

btks
The BTK/TEC-family kinases are enzymes that phosphorylate tyrosine residues.They belong to the non-receptor or cytoplasmic protein tyrosine kinase (PTK) superfamily.The non-receptor PTK proteins are localized in the cytoplasm entirely and interact with the cytoplasmic portions of cell surface receptors. In mammals theBTK/TEC kinases play important roles in the proliferation and differentiation of cells of the adaptive immune system. In mammals, the BTK/TEC gene family is comprised of five paralogous genes: a) TEC,b) BTK, c) BMX, d) ITK, and e) TXK.
We explored the origin, diversification, and functional differentiation of the well-studied btk/tec family of kinase genes. Interestingly, in mammals the N‐ and C‐terminal domains of these kinases— PH and SH1 respectively—are conserved. In contrast, the SH3 and SH2 domains and their flanking linker sequences (SSL region) are highly diverged. However, the SSL region of paralogous BTK/TEC proteins has not differentiated with respect to its protein ligands. Therefore we hypothesized that this region has differentiated with respect to a different function, that of lipid binding. To test this hypothesis, we determined that the SSL region has the capacity to bind to lipids, and second verified the functional differentiation hypothesis by showing that the SSL region of paralogous btk/tec genes binds to different lipids. We also showed that natural mutations in the SSL region of the btk gene, which cause a hereditary immunodeficiency in mice and humans, alter the lipid‐binding profile of the BTK protein. Our study revealed a new function for the BTK/TEC kinases. It also suggested a link betweenthe lipid‐binding function and the regulation/function of B cells.
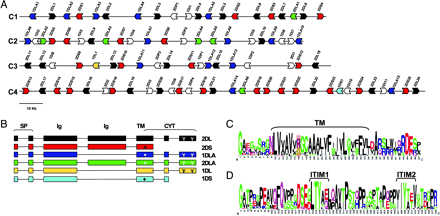
chirs
In mammals, the cell surface receptors encoded by the leukocyte receptor complex (LRC) regulate the activity of T and B lymphocytes, as well as that of natural killer cells, and thus provide protection against pathogens and parasites. Contrary to the generally accepted view, we showed that immunoglobulin‐type receptors functioning in the innate immune system are present in the non‐mammalian genomes.
Specifically, we found that the chicken genome encodes many Ig-like receptors that are homologous to the LRC receptors, and showed that the chicken Ig-like receptor (CHIR) genes are members of a large monophyletic gene family. The members of this monophyletic family are organized into genomic clusters, which are in conserved synteny with the mammalian LRC. One-third of CHIR genes encode polypeptide molecules that contain both activating and inhibitory motifs. These genes are present in different phylogenetic groups, suggesting that the primordial CHIR gene could have encoded both types of motifs in a single molecule. In contrast to the mammalian LRC genes, the CHIR genes with similar function (inhibition or activation) are evolutionarily closely related. We proposed that, in addition to recombination, single nucleotide substitutions played an important role in the generation of receptors with different functions. Structural models and amino acid analyses of the CHIR proteins revealed the presence of different types of Ig-like domains in the same phylogenetic groups, as well as sharing of conserved residues and conserved changes of residues between different CHIR groups and between CHIRs and LRCs. Our data supported the notion that the CHIR gene clusters are regions homologous to the mammalian LRC gene cluster, and favored a model of evolution by repeated processes of birth and death (expansion–contraction) of the Ig-like receptor genes.